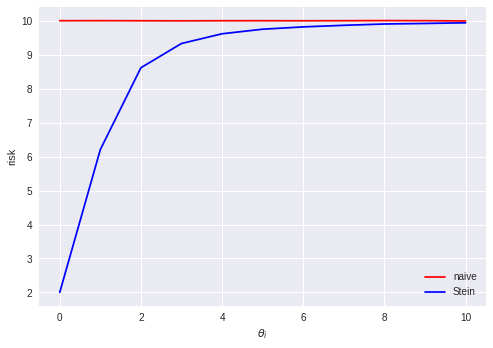
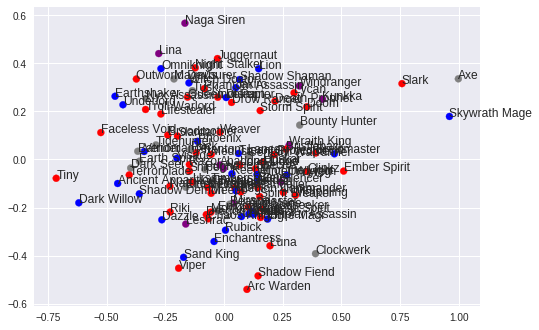
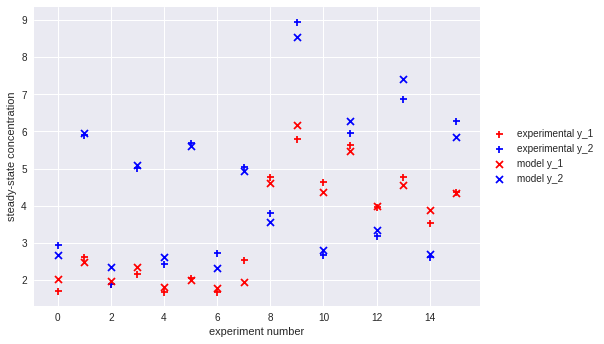
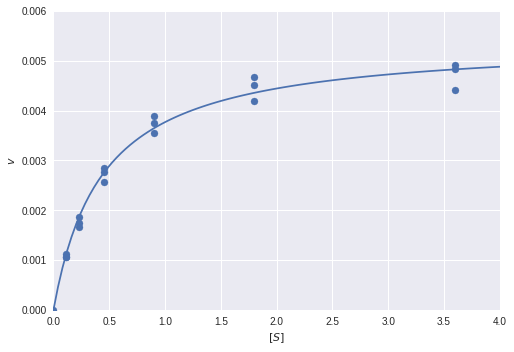
Linear regression in Python (UPenn ENM 375 guest lecture)
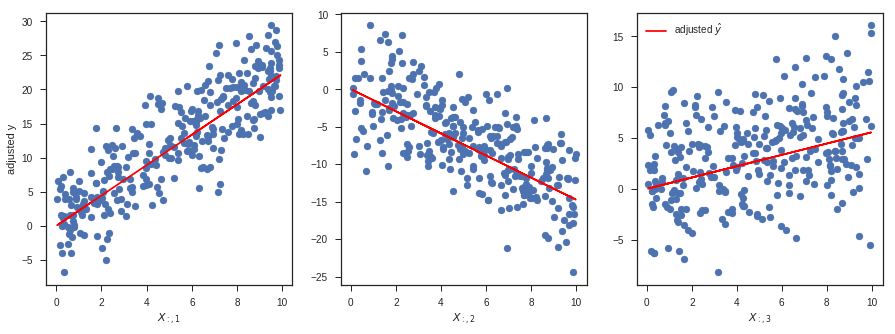
I was recently invited to give a guest lecture in the course ENM 375 Biological Data Science I - Fundamentals of Biostatistics at the University of Pennsylvania on the topic of linear regression in Python. As part of my lecture, I walked through this notebook. It might serve as a useful reference, covering everything from simulation and fitting to a wide variety of diagnostics. The walkthrough includes explanations of how to do everything in vanilla numpy/scipy, scikit-learn, and statsmodels. As a bonus, there's even a section on logistic regression at the end.
Read on for more!